RPcontact: Improved prediction of RNA-protein contacts using RNA and protein language models
RPcontact: Improved prediction of RNA-protein contacts using RNA and protein language models
Jiang, J.; Zhang, X.; Zhan, J.; Miao, Z.; Zhou, Y.
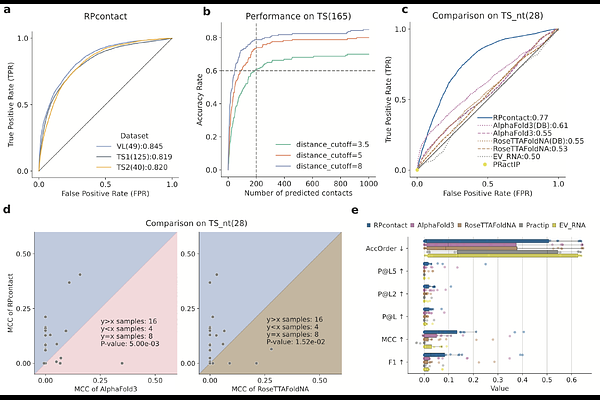
AbstractDetermining key contacts between RNA-protein interactions is essential for understanding the molecular mechanisms of numerous biological processes, including transcription, splicing, and translation. However, progress in this area has been impeded by the scarcity of RNA-protein complex structures in the Protein Data Bank (PDB) and the challenges posed by traditional structural determination techniques. Recent computational advancements, including deep learning methods like AlphaFold 3 and RoseTTAFoldNA, have improved contact prediction but are still limited by the availability of homologous sequences and templates. Here, we introduce RPcontact, a novel computational method designed to predict RNA-protein contacts using large language models tailored for RNA (ERNIE-RNA) and proteins (ESM-2). Despite being trained entirely on ribosomal RNA-protein (rRNA-protein) complexes, RPcontact demonstrates robust and generalized performance in predicting contacts for both dimeric and multimeric non-rRNA-protein complexes. The performance of RPcontact on contact predictions significantly improves over the binary contacts inferred from RNA-protein complex structures predicted by AlphaFold 3 and RoseTTAFoldNA, highlighting its potential in RNA-protein complex structure and function prediction.