Chemical Probe Discovery for DEAD-box RNA Binding Protein DDX21 using Small Molecule Microarrays
Chemical Probe Discovery for DEAD-box RNA Binding Protein DDX21 using Small Molecule Microarrays
Calo, E.; Koehler, A. N.; Aiba, T.
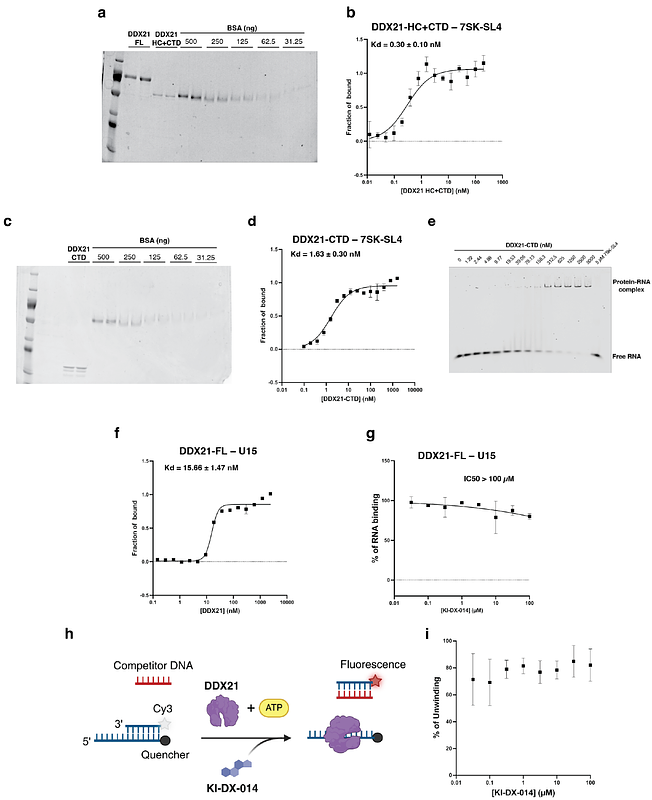
AbstractThe DEAD-box family of ATPases plays a critical role in nearly all stages of RNA metabolism, from transcription to degradation, and serves as a major regulator of biomolecular condensates. Dysregulation of DEAD-box proteins is well established in a variety of diseases, including cancer and neurodegenerative disorders, making them attractive therapeutic targets. However, their classification as \"undruggable\" has historically hindered small molecule-based modulation. In this study, we focus on DDX21, a member of the DEAD-box family involved in ribosome biogenesis and transcriptional regulation. As a proof of concept for targeting such RNA-binding proteins, we developed a lysate-based small molecule microarray platform to identify compounds that directly bind DDX21. This screen led to the discovery of KI-DX-014, a small molecule compound capable of inhibiting DDX21 interaction with RNA. KI-DX-014 modulated RNA-dependent functions of DDX21, including its ATPase activity and biomolecular condensate formation. Furthermore, KI-DX-014 attenuated the DDX21-dependent release of P-TEFb from the 7SK snRNP complex in vitro, suppressed P-TEFb-dependent phosphorylation of the RNA polymerase II CTD, and induced developmental defects in zebrafish embryos. These findings reveal a previously unexploited therapeutic avenue and establish KI-DX-014 as a valuable chemical probe for dissecting the biological functions of DDX21 in both normal physiology and disease states.