GeneChat: A Multi-Modal Large Language Model for Gene Function Prediction
GeneChat: A Multi-Modal Large Language Model for Gene Function Prediction
Dhanasekar, S.; Saranathan, A.; Xie, P.
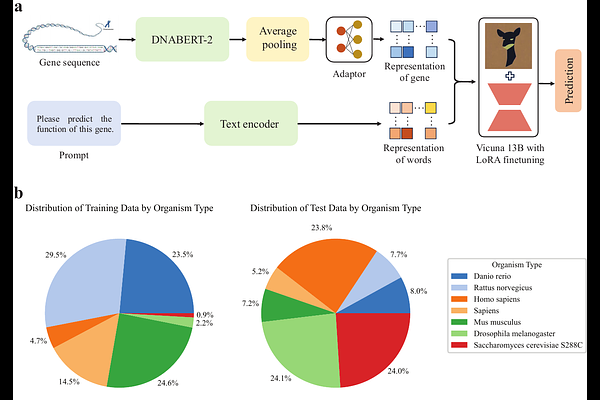
AbstractAccurately predicting gene function from DNA sequences remains a fundamental challenge in genomics, particularly given the limited experimental annotation available for most genes. Existing computational approaches often formulate function prediction as a classification task over predefined categories, limiting their flexibility and expressiveness. We introduce GeneChat, a multi-modal large language model designed to generate free-form, natural language descriptions of gene functions directly from nucleotide sequences and textual prompts. GeneChat integrates three components: a DNABERT-2-based gene encoder optimized for long-range genomic context, an adaptor that aligns gene representations with the input space of a large language model, and Vicuna-13B, a fine-tuned LLaMA-2 variant used to produce coherent functional narratives. Trained on over 50,000 genes from the NCBI database, GeneChat outperforms GPT-4o on BLEU and METEOR metrics, demonstrating superior ability to generate accurate, context-aware, and semantically rich descriptions. This work highlights the potential of foundation models for advancing interpretable and scalable gene function prediction in a free-form, language-driven paradigm.