Molecular Phenotypic Plasticity Informs Possible Adaptive Change of Triple-Negative Breast Cancer Cells In Vivo
Molecular Phenotypic Plasticity Informs Possible Adaptive Change of Triple-Negative Breast Cancer Cells In Vivo
Iftehimul, M.; Muganda, P. M.; Newman, R. H.; Rorie, C. J.; Harrison, S. H.; Hossain, M. T.; Thomas, M. D.; Graves, J. L.; Saha, D.
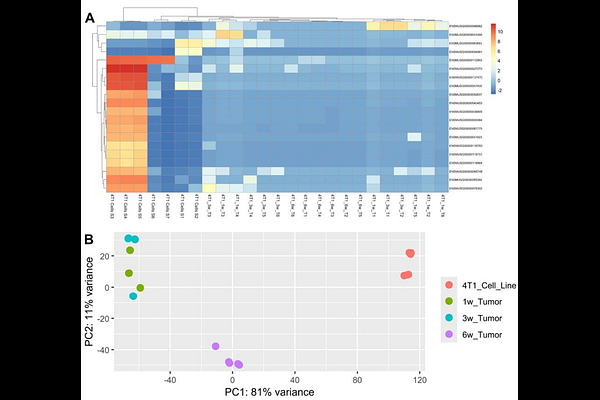
AbstractBackground and Objectives: Cancer evolves via interconnected mechanisms, including changes in extrachromosomal DNA (ecDNA), genetic instability, and interactions with the tumor microenvironment (TME). These mechanisms allow for some clones to evolve metastatic traits, evade the immune system, and resist chemotherapy. However, how cancer cells evolve in vivo remains poorly understood. This study investigates the in vivo changes in gene expression of triple-negative breast cancer (TNBC) cells implanted in BALB/c mice. Methodology: We analyzed RNA-seq data from 4T1 TNBC cells and tumors at different growth stages (1-, 3-, and 6-week) to identify differentially expressed genes, protein-protein interactions, and ecDNA alterations. We also assessed how ecDNA and genomic instability proteins interact with anti-TNBC drugs. Results: Our results reveal early transcriptional shifts within one week of tumor implantation, showing rapid acclimation. Changes in gene expression continued over time, with significant molecular reprogramming observed at six weeks under in vivo environmental pressures, including ecDNA alterations and immune evasion. The shift from the earlier generation (1 week) to the later generation (6 weeks) suggests cumulative alterations in key oncogenic pathways related to tumor progression. Additionally, we found that mutations in ecDNA and genomic instability proteins influence drug binding affinity, suggesting that adaptive changes may impact chemotherapy response. Conclusions and Implications: This study provides new insights into how TNBC tumors may evolve over time and novel ecDNA-related mechanisms of possible tumor adaptation, highlighting potential biomarkers for tumor aggression and immune evasion, which could help develop more effective therapeutic strategies against TNBC.