A community-consensus reconstruction of Chinese Hamster metabolism enables structural systems biology analyses to decipher metabolic rewiring in lactate-free CHO cells
A community-consensus reconstruction of Chinese Hamster metabolism enables structural systems biology analyses to decipher metabolic rewiring in lactate-free CHO cells
Di Giusto, P.; Choi, D.-H.; Antonakoudis, A.; Duraikannan, V. G.; Craveur, P.; Cowie, N. L.; Ganapathy, T.; Ramesh, K.; Benavidez-Lopez, S.; Orellana, C. A.; Jimenez, N. E.; Dworkin, L. A.; Morrissey, J.; Marin de Mas, I.; Strain, B.; Valdez-Cruz, N. A.; Trujillo-Roldan, M. A.; Marzluf, J.; Martinez, V. S.; Zehetner, L.; Altamirano, C.; Vega-Letter, A. M.; Priem, B.; Cao, H. C.; Hold, M.; Ma, J.; Hong, Y.; Gopalakrishnan, S.; Enuh, B. M.; Tarzi, C.; Pang, K. T.; Angione, C.; Zanghellini, J.; Kontoravdi, C.; Hefzi, H.; Betenbaugh, M. J.; Nielsen, L. K.; Lakshmanan, M.; Lee, D.-Y.; Richelle, A.;
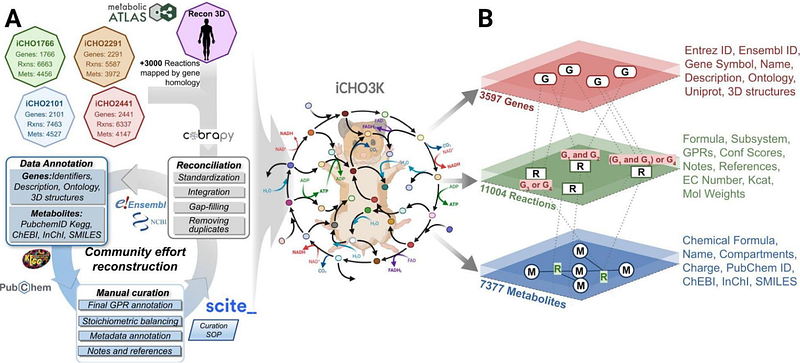
AbstractGenome-scale metabolic models (GEMs) are indispensable for studying and engineering cellular metabolism. Here, we present iCHO3K, a community-consensus, manually-curated reconstruction of the Chinese Hamster metabolic network. In addition to accounting for 11004 reactions associated with 3597 genes, iCHO3K includes 3489 protein structures and structural descriptors for >70% of its 7377 metabolites, enabling deeper exploration of the link between molecular structure and cellular metabolism. We used iCHO3K to contextualize transcriptomics and metabolomics data from a CHO cell line in which lactate secretion is abolished. We found the reduced glycolytic flux and enhanced TCA cycle flux were accompanied by an elevated NADH and PEP levels in these cells, consistent with experimental measurements. Leveraging iCHO3Ks structural annotations, we identified candidate binding interactions of NADH and PEP with glycolytic enzymes showing model-predicted differential flux, suggesting novel allosteric regulation associated with the observed decrease in glucose uptake and glycolysis. Overall, iCHO3K offers a valuable framework for systematic integration of omics data, improved flux predictions, and structure-guided insights, thus advancing CHO cell engineering and enhancing biomanufacturing efficiency.