Analysis of Divergent Gene Expression between HPV+ and HPV- Head and Neck Squamous Cell Carcinoma Patients
Analysis of Divergent Gene Expression between HPV+ and HPV- Head and Neck Squamous Cell Carcinoma Patients
Shankar, K.; Walker, S. E.
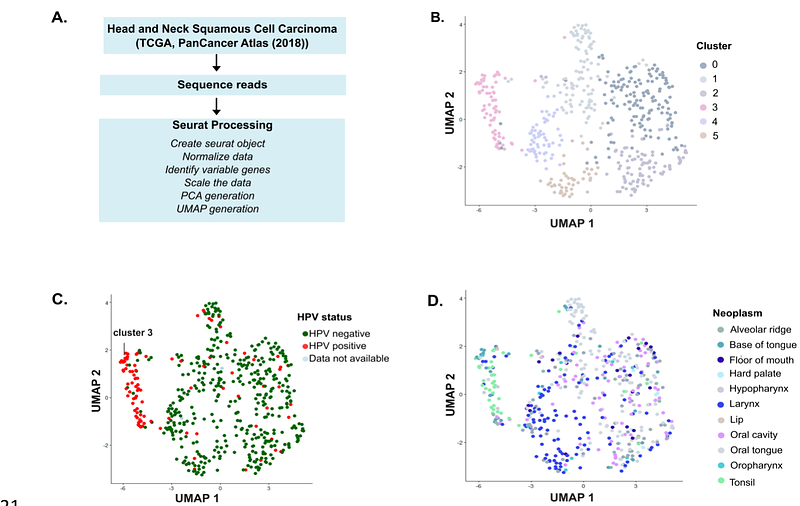
AbstractHuman Papillomavirus (HPV) is a non-enveloped virus with a circular double-stranded DNA genome. It is one of the most common sexually transmitted infections, with high-risk types such as HPV-16 and HPV-18 linked to anogenital and head and neck squamous cell carcinomas (HNSCC). HNSCC includes cancers of the oral cavity, pharynx, larynx, and related regions, caused by carcinogens or persistent viral infections. HPV-positive (HPV+) HNSCC cases are more prevalent in Western countries and exhibit better prognosis and treatment response compared to HPV-negative (HPV-) cases. These differences suggest distinct fundamental differences between each subtype. This study analyzed RNA-seq data from the PanCancer Atlas 2018 dataset to investigate molecular distinctions between HPV+ and HPV- HNSCC. Using dimensionality reduction techniques such as Principal Component Analysis (PCA) and Uniform Manifold Approximation and Projection (UMAP), a clear clustering of HPV+ cases was observed, suggesting a unique gene expression profile. HPV+ tumors exhibited upregulation of genes involved in nucleic acid processing and downregulation of genes associated with apoptosis and epidermis development. These findings underscore the biological differences between HPV+ and HPV- HNSCC, offering insights into HPV-driven oncogenesis. Understanding these distinctions may improve patient stratification and inform targeted therapeutic strategies for HNSCC.